[Course site][IISER-p]
Bio322: Biophysics I-
[ver.2014]
Physical biology of molecules, cells and tissues: Equilibrium
approaches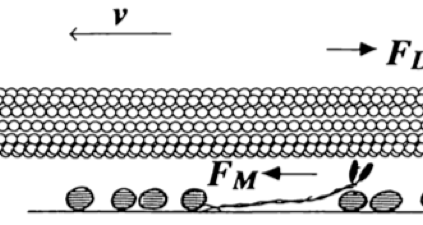
S. No. |
Title |
Due date |
Form of submission |
Date posted |
Additional Information |
Solution |
| 1 |
Dimensions and orders of
magnitude estimates |
25-Aug-2014
@1800h [NOTE: time postponed by 10h] |
Upload on course website
1) iiser-TVM [FOLDER]
(no login required), or send an email to: bio322 at the rate students
dot iiserpune dot ac dot in. 2) iiser-PUN: hard copies
only.
|
18-Aug-2014 |
Total score: 16 pts
[Please upload/send ONLY PDF files]
|
Solution to dimensions and
OOME (updated with some changes to the dimensions answer) |
| 2 |
Calculating the radius of gyration of
proteins from their 3D atomic structures |
29-Oct-2014 @0800h [AGAIN postponed
from previous date]
|
Form
for upload (bugs fixed w.r.t. access control 2014/10/20) |
17-Oct-2014 |
Octave demo in class on 20-Oct-2014
Rasmol
Pymol
Rasmol
FAQ:
Measure atom-atom distances:
Settings->(set picking distance)
Mouse-click (single click) at two points in space
Dist. in Angstrom reported in display and command-line.F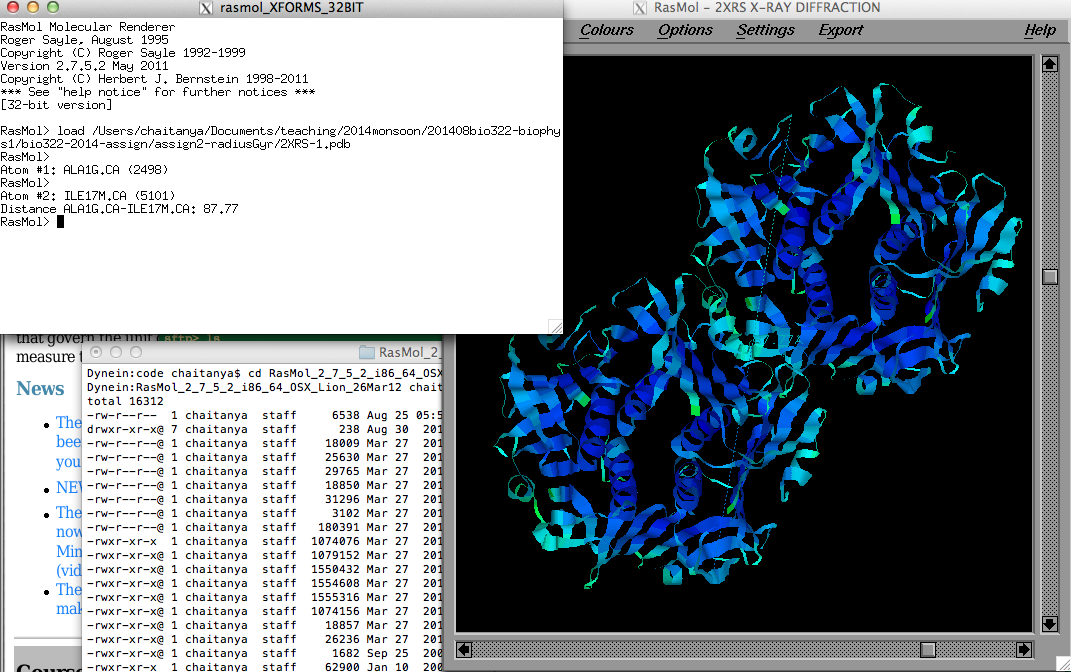
[read_pdb.m]
[GFP]
[NS1]
[LacI]
|
|
| 3 |
Persistence length |
20-Nov (in classs) |
Explanation and solved in class |
18-Nov-2014 |
Calculate the persistence length of
MTs
MT-image
|
|
Teaching assistant (TA)
Names: 1. Neha Khetan (PhD student, IISER Pune), 2.
Kunalika Jain (PhD student, IISER Pune), 3. Sreejith A. (PhD student, IISER
Thiruvananthapuram)
Where to find the TAs: 1. C203, hr4, IISER Pune. 2: G1, 1st
lab, right side, IISER Pune, 3: IISER Tvm.
Instructions for submisions
- Assignments to be submitted either in hand-written form or electronic
file in PDF format (or hand-written), as per conditions specified.
- IF electronic submissions are being accepted, please send them to the
email address bio322 at the rate
students dot iiserpune dot ac dot i
n. In the email please include the following subject
line: "Assignment x- y" where x is the assignment serial number (1,2,3...)
and y is the name of the assignment (eg.: Olympics of organisms).
- The file name should be "Axx_name_.pdf"
where xx is the two-digit assignment number 01, 02, etc.
- I prefer PDF submissions from LaTex source. Word doc or Open Document
Format (ODF) submissions must first be converted to PDF before submission.
Sometimes DOC->PDF conversions are platform- if all else fails, I will
accept a DOC or ODF file (i.e. do not send docx files).
- Submissions will be checked for copy-pasting.
- If hand-written submissions are expected, please ensur
your own name and roll number are written on top on page 1, ensure
legibility and make a clear indication which question or section is being
answered.
- Please do not hand in submissions to me in corridors,
staircases, the canteen or other locations. It is liable to be
lost. It is your responsibility to hand it to
me or to the TA in our lab/office.
Updated: 18 Nov 2014

