Emergence of spontaneous rotation in asters confined in a 2D circular boundary.
Cytoskeleton and Cell Shape (CyCelS) Lab
We are curious to answer the question how when more than one molecular motors act on microtubule (MT) cytoskeleton, what are the collective forces generated. To this end we work with purified moecular motors in in vitro reconstitutions and observe our system under microscopes. The collective effects that we observe of motor numbers, filament lengths and velocities of transport are then used to infer how these processes may drive cell shape, polarization and division. We also use computational models- equations solved in numerical simulations – to compare our expections with measurements from experiments. As a cellular model we have adopted the rod-shaped cell E. coli to examine the role of actin- and microtubule-like proteins in their size and shape regulation. This cycle of experiment<->theory<->comparison<->model refinement is our discovery cycle.
Modeling the mechanobiological constraints on mitotic spindle oscillations in nematodes across evolution

Cytoplasmic viscosity estimates from yolk granule mobility. The figure is taken from the paper by Khatri et al. Mol Biol. Cell
The first embryonic division of the well studied worm Caenorhabditis elegans shows asymmetric division- one cell is smaller (posterior) and the other bigger (anterior). The evolutionary diversification of related embryonic divisions is an ideal framework to study rules driving divergence in collective outcomes- in this case spindle oscillations. In collaboration with the lab of Prof. Marie Delattre, Plasticity and Evolution of Cell Division Lab at ENS Lyon, we have studied the problem. And find a new biophysical parameter that could predict the onset of oscillations in yet unseen species.
Lattice-based diffusion and aggregation simulations: from receptors to microtubules
Receptor aggregation dynamics by dimerization is thought to be critical for signalling in cancer cells, as summarized in a theoretical model by us in 2005 (Athale, Mansury & Deisboeck J. Theor. Biol.). The microscopic details of such receptor dynamics have been more recently modeled using coarse-grained simulations in collaboration with the Biophysical Chemistry lab in NCL Pune (Pawar et al. 2014, Sengupta et al. 2016). We have more recently developed a Monte-Carlo simulation of receptor dimerization kinetics based on a diffusion and aggregation model in a heterogenous environment (Deshpande et al. (2017) Phys. Biol).
Simulating collective microtubule-motor Transport: Modelling mechanical cooperativity in microtubule transport in spindles, neurons and flagella
In recent work we have modelled the self-organized pattern formation of multiple microtubule (MT) asters by the interaction of the MTs with kinesin-5 like molecular motors. In an iterative manner we have demonstrated that the structures surprisingly follow the statistics of “circle packing” seen in classical geometrical systems. This problem has its origins in Gauss and Lagrange’s work on optimal packing patterns. You can read more in the Journal of Cell Science (Khetan, et al. 20021).

Simulating aster tessellation of oocyte space: Model of Phallusia intracellular microtubule patterns
Experimental collaborator Dr. Janet Chenevert, Celine Hebras and Dr. Gerard Pruliere, Obs. Oceanongraphique, Villefranche sur Mer, France
In vitro reconstitution of purified molecular motors and microtubules (MTs)
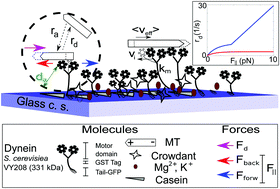
EXPT: Collective transport by molecular motors of teams: Pattern formation in vitro
Fluorescently labelled Mts transported by a sheet of molecular motors
When single-molecule mechanics of motors is studied it reveals a lot about the mechanical properties. We seek to try to take these measurements and put them “together” in terms of a collective transport system, in vitro. To obtain inferences, we combine it with a computational model based on detailed single-molecule mechanics (Jain, Khetan and Athale 2019) integrates experiment and theory. This approach has led us to discover a jump between 5-10 motors per MT resulting in directional transport by yeast cytoplasmic dynein, using the minimal construct developed in Prof. Ron Vale’s lab (Reck-Peterson et al. Cell doi: 10.1016/j.cell.2006.05.046).
Aster positioning in small and large cells:
Ex vivo Xenopus extract experiments (Athale et al. 2014 Phys. Biol. 11: 016008) and the in vivo self-organisation of spindles in mouse oocyes undergoing meiosis I (Khetan & Athale, 2016 PLoS Comp Biol) were both modelled using a gliding assay with asters.
Self-organized spontaneous rotation of MT asters
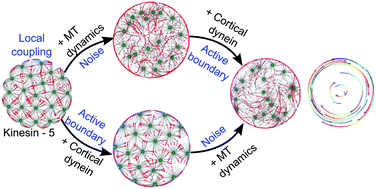
Models of locally coupled self-propelled particles (SPPs) that can self-organise to spontaneously undergo non-chiral rotations
When we further explored the pattern forming nature of the simulated asters, in cells of larger and smaller sizes, we discovered to our surprise that the asters interacting with a reflecting boundary, dynein at the cortex and EG5 like activity in the cytoplasm (kinesins that bind to pairs of asters and couple them to either zipper or separate them) can generate these patterns (Khetan and Athale, 2020, Soft Matter).

Self-organized swarming of MT asters
Nuclear positioning by dynein in mitosis
In more recent work we have begun to examine how these number-dependent effects affect the transport and positioning of nuclei in the budding yeast S. cerevisiae.
Mathematical model of microtubule dynamics in neuronal growth cone turning
In order to examine role of the MT system in axonal growth cones and their limit to direction-sensing we developed a model combining reaction-diffusion with stochastic microtubule-motor mechanics and effective actin flows driven by myosin. The model predicted limits to directional cues. (Mahajan and Athale 2012).
Tubulin polymerization kinetics and dynamics: Kinetic modelling reconciled to experiment

(Jain et al. 2022 Cytoskeleton) Simulating the kinetic competition between microtubule (MT) nucleation and GTP hydrolysis.
The diversity of microtubule (MT) inhibitors that originates from plants and have medicinal uses has fascinated us. begun to investigate the role of MT polymerization kinetics in presence of colchicine – both produced from the plant and present in the insect that feeds on it.
Quantitative Microscopy: Imaging Analysis and BigData Analytics
Increasingly we find the large datasets that were previously restricted to DNA, RNA and Protein data (genomics, transcriptomics and proteomics) have spread to image-data. These coming from High Content Screening (HCS) for genetic and drug discovery have the potential to “discover” not just sequence and structure, but functional relations between proteins in the context of spindle assembly and cell polarization. Our ambitions in this part of our lab is to build data pipelines to feed into our theoretical models and even use AI tools to Discover biology from the data alone. Some new work in the lab in collaboration with an Evolutionary Developmental Biologist (Dr. Marie Delattre, ENS Lyon, France) is in preliminary stages. Watch this stage for more.

The GUI of the DIC object tracking (DICOT) program. This is part of a publication by Chaphalkar*, Jawale*, Khatri and CAA 2021 Biophy J. (* equal contribution)
Mechanics of bacterial cell size and growth: Connecting cytoskeleton to cell shape in simple rod-shaped bacterial cells

Simulations and experimental data of scaling of E. coli grown on agarose pads are compared to estimate the SA-V scaling exponent.
The cell size variability in cell shape in a population (Athale &Chaudhari Bioinformatics) is thought to be linked to replication processivity linking the rate of growth of single cells to cell division (Gangan & Athale 2017 Roy. Soc. open science). We are currently working on using the variability to re-visit allometry relations (Kale et al. 2023). The scaling behaviour of surface area (SA) ∝ volume (V) that deviates from γ~ 2/3 (geometric scaling law) points to active cellular mechanics and regulation driving the scaling. We are using this insight to look top-down using experiments and simulations to get a picture of the mechanical role of cytoskeleton in regulating cell shape in rod-shaped bacteria.

The variability in a population of growing E. coli on an agar-pad
Philosophy of the lab: Collective pattern formation (Eka, Aneka aur Ekta)
As with flocks of birds, schooling of fish and people milling at a railway station, collective patterns that emerge have fascinated humankind since time immemorial. We hope to work on biomolecules and find out the laws by which they organize. Finding the similarities and differences to more complex pattern, is our way of working our way upwards in complexity, in the hope of finding out the universal laws and the exceptions that guide such patterns. The 1974 parable from Films Division India “एक, अनेक और एकता”, a must watch.

Collective effects self-organized: in a social context (Films Division India): This social parable from Films Division India (1974) has amazing parallels in collective behaviour as we think of it- birds swarming, people at a railway station, fish schooling.
Funding
- Dept. of Biotechnology, Govt. of India- Bioinformatics Centre with labs of Dr. MS Madhusudhan & Dr. Saikrishnan K.
- Shastri Indo-Canadian Institute
- Indo French Centre for the Promotion of Advanced Research (CEFIPRA)

CEFIPRA: Indo French Agency for Promotion of Advanced Research
- Department of Science and Technology, Govt. of India

- Private companies for some short term projects with BS-MS undergraduate students (iGEM)