How to use HIstome: The Histone Infobase
The Histone Infobase is an online database that provides detailed information about human histones, their post-translational modifications and enzymes responsible for addition and removal of these modifications.
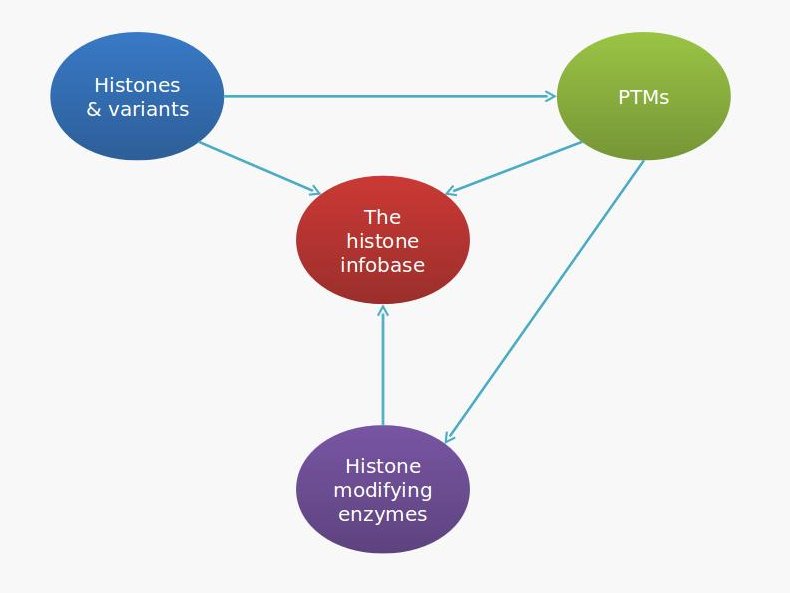
Figure 1: Modules of the histone infobase.
For ease, the database has been divided into following sections which can be browsed through the side-menu.
Lead-in: This section provides basic information about histone protein family, and its importance in chromatin compaction and gene regulation within the cell. The section also introduces the user to post-translational modifications(PTMs) of histone proteins and their significance in cellular processes and gives a glimpse of plethora of enzymes that carry out these modifications.
The database is futher divided into four browsable sections as follows...
Histones and histone variants: This section takes the reader one step ahead into the diversity of histone proteins. It is divided into subsections that stand for different types of histones. Each subsection takes the reader deeper into protein diversity within that histone-type by providing a table of distinct proteins in that category. A provision has been made to download this table as an excel file. The page also provides a link to NCBI MapViewer that displays the chromosomal map of all genes coding for histone proteins in that category.
Each histone entry in the table opens into its own page that provides manually curated information about that protein. The page also provides a table of protein, mRNA or gene related accessions that connect the user to related databases. The page provides upstream sequence of each gene for promoter analysis and also tabulates information about associated diseases, if any. The page additionally provides a graphical interface for sites of post-translational modifications of that histone (Figure 2). Each site of modification, a combination of a single letter amino acid code and a double letter modification code provides a hyperlink to its own separate page. This histone modification page can also be reached by browsing through 'Histone PTM' section as descibed further.
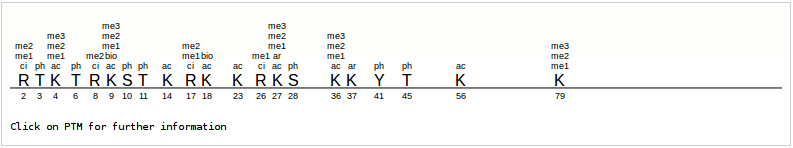
Figure 2: Graphical representation of modification code. Example: Histone H3.3
Histone PTMs: This section provides the user with details about known histone post-translational modifications. The section is divided into subsections that stand for different types of modifications. Each subsection opens into a separate page that diplays a list of different sites of modifications in that category. Each site of modification has been hyperlinked to its own separate page that provides manually curated information from literature and a table of enzymes that 'write' or 'erase' these modifications from the histones (Figure 3). Each enzyme has in-turn been hyperlinked to its individual page that can also be reached by browsing the 'The writers' and 'The erasers' sections of the menu as described further.
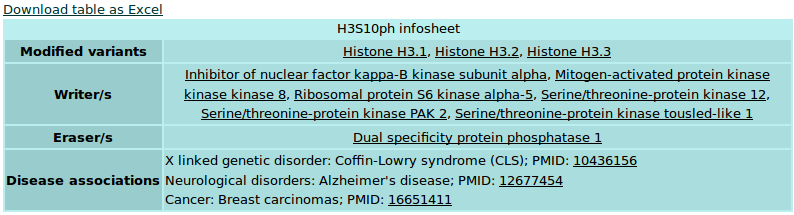
Figure 3: Table for post-translational modification. Example: Histone H3 Serine 10 phosphorylation
Histone modifying enzymes have been divided into two categories, 'writers' that add and 'erasers' that remove modifications from histones. Though, further information about writers and erasers has been provided in two different sections, the structure of both the sections remains logically same.
Both writers and erasers have been divided into subsections depending upon the type of modification and the type of amino acid modified by them. Each subsection provides a list of enzymes that fall into that category (Figure 4). The page also provides a link to NCBI MapViewer to view chromosomal map of genes coding for enzymes that belong to the particular category. Each enzyme has been hyperlinked to its own page that displays manually curated information from literature. The enzyme page also provides a tabulation of protein, mRNA or gene related accessions that connect user to other related databases and disease related information
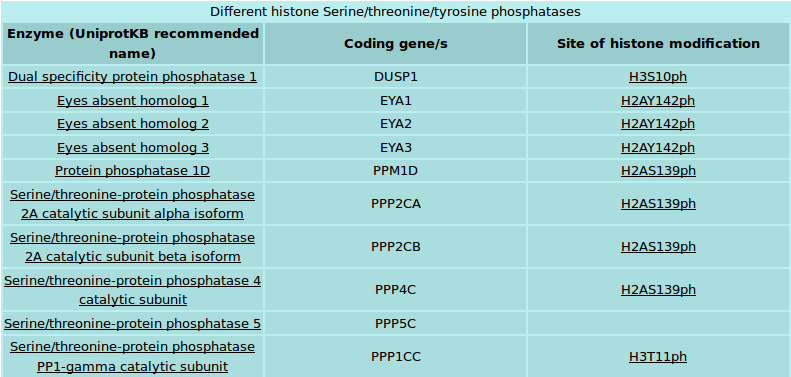
Figure 4: Table of histone modifying enzymes. Example: Serine/threonine/tyrosine phosphatases
Search HIstome: The database contents can be searched by either the Google powered search or the HIstome advanced search. The Google powered search lists all the pages containing the search term as ranked by Google. The Histome advanved search on the other hand allows user to search terms in specific categories such as histones, enzymes, PTMs or diseases. The advanced search options allows user to retrieve results that can neither be browsed nor searched for using Google powered search. Such results include list of all PTMs of paricular type of histone, list of enzymes involved in certain type of disease etc.