Lead-in
Size of the haploid human genome is about 2.9 billion base pairs. When DNA from a single diploid eukaryotic cell in the 23 pairs of chromosomes is attached end-to-end, its total length of DNA measures about 2 meters. On the other hand, the diameter of a typical eukaryotic nucleus is roughly 6 micrometer. The tiny volume of the nucleus therefore raises an important question as to how is the complete genome accommodated in such a confined space without making any changes in the size or functional attributes of the genome? Thus, this poses a 'packaging problem' for the cellular machinery. What sort of a design can facilitate the packaging of DNA, create a dynamic architecture that also provides space for enzymatic machineries of transcription and replication and at the same time maintain the three-dimensional organization of genomic loci such that they have a non-random location in the nucleus?
Unbelievably, the answer lies in the magnificent structure of Chromatin.
What is Chromatin?
Low molecular weight, highly basic proteins called 'Histones' associate with DNA in a definite stoichiometry to constitute the fundamental repeating unit called 'Nucleosome' that forms the basis of Chromatin. Genomic DNA wraps around the histone octamers at specific intervals resulting in the formation of a beads-on-a-string like structure called as the '10 nm fiber' that provides roughly 6-fold unit compaction. This fiber is then further condensed into a series of hierachical structures that cause further compaction of the genome. The final step in the compaction process yields a structure that is almost 10,000-fold more compact as compared to the original and forms the peculiar structures called Chromosomes. Formation of a chromosome is essential for the unperturbed and symmetric distribution of the genomic content between the two progeny that form after cell division. Such a structure also provides a sturdy design that can bear the physical stress and strain conferred upon it by the pulling forces of the spindle fibers. After cell division, the compact chromosomes undergo series of decompaction events. Thus, the hallmark of the hierachical organization of chromatin higher-order structures is that they are inter-convertible depending upon the requirement of a cell.
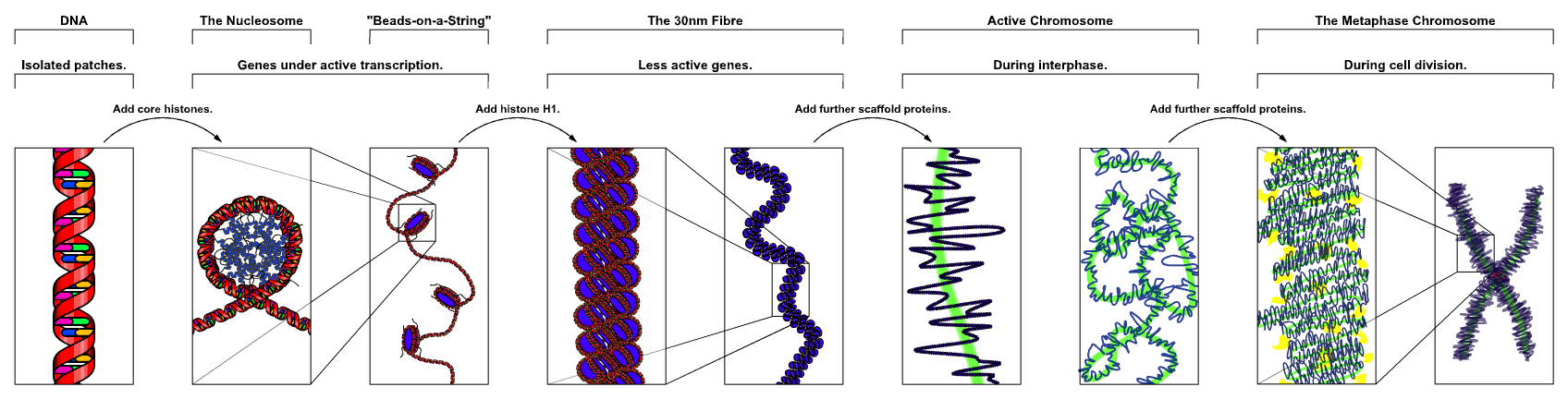
Figure: Major chromatin structures.
What are Histones?
Histones are small molecular weight proteins ranging 14 to 18 kDa in size and are enriched in basic amino acids. There are 5 different types of histones: H1, H2a, H2b, H3 and H4. Histones are ubiquitous and among the most conserved proteins from yeasts to humans. Each type of histone has its own subset of variants that have been shown to play a significant role in the formation of epigenetic memory and many different cellular processes. Post-translational modifications of these histone/histone variants by 'histone modifiers' paves the way for epigenetic mechanisms of gene regulation.
What is a Nucleosome?
Dimers of Histones H2a-H2b and H3-H4 combine to form an octamer that wraps around 146 base pair of DNA in ~1.6 turns to comprise a structure called as nucleosome. The positively charged residues in Histones form tight complex with the negatively charged DNA by virtue of electrostatic forces.
What are histone modifiers?
Studies in recent years have identified plethora of enzymes that mediate the addition of various small chemical groups such as acetyl, methyl, phosphate, ubiquitin etc. onto the N-terminal tails of histones. Such enzymes are called as 'writers' since they create specific epigenetic marks that contribute in alteration of the gene expression profile of that locus and/or towards alteration in chromatin architecture. On the other hand, the enzymes that remove such marks from the chromatin template so as to allow a restructuring of the expression state or chromatin architecture of a locus are categorised as 'erasers'.