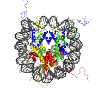
Histone-lysine N-methyltransferase EZH2
Ezh2 is part of Polycomb group of repressor complex (PRC) that are crucial for the silencing of Hox genes by the H3K27 methylation activity of Ezh2. PRC2 is also involved in the methylation and inactivation of X chromosome. H3K27 methylation by ezh2 is also required for maintenance of pluripotency (PMID: 15196462). Ezh2 is overexpressed at mRNA as well as protein level in many types of cancers like breast cancer, prostate cancer and are related to the aggressiveness and progression of these carcinomas (PMID: 12374981, PMID: 14500907). It is also shown to downregulate tumour suppressor genes like E-cadherins via H3K27 methylation (PMID: 18806826).
| UniprotKB Accession | Q15910 | |
|---|---|---|
| Alternative Name(s) | ENX-1, Enhancer of zeste homolog 2, Lysine N-methyltransferase 6 | |
| Writer for PTM/s | H3K27me1, H3K27me2, H3K27me3, H1K25me1 | |
| EC number | 2.1.1.43 | |
| No. of coding genes | Histone-lysine N-methyltransferase EZH2 is coded by following 1 non-allelic gene/s | |
| Gene name | enhancer of zeste homolog 2 (Drosophila) | |
| Gene symbol | EZH2 | |
| Promoter region (-700 TSS +300) | Get sequence | |
| Previous Symbol/s | ||
| Aliases | EZH1, ENX-1, KMT6, KMT6A | |
| GeneID | 2146 | |
| Chromosomal location | 7q35-q36 | |
| HGNC | 3527 | |
| Unigene | Hs.444082 | |
| MIM | 601573 | |
| RefSeq mRNA | NM_004456.4 | NM_152998.2 |
| RefSeq Protein | NP_004447.2 | NP_694543.1 |
| Disease associations | Cancer: Metastatic prostrate cancer; PMID: 12374981 Cancer: Humanmantle cell lymphoma; PMID: 11298590 Cancer: Breast cancer; PMID: 11389032 Cancer: Hodgkin's lymphoma; PMID: 14500907 Cancer: Renal cell carcinoma; PMID: 20920340 Hematological disorder: Myelodysplastic syndromes (MDS); PMID: 20601954 Cancer: Cisplatin-resistant ovarian cancer cells; PMID: 20686362 | |
Lysine methyltransferases Histone-lysine N-methyltransferase EZH1, Histone-lysine N-methyltransferase EZH2, Histone-lysine N-methyltransferase MLL, Histone-lysine N-methyltransferase MLL2, Histone-lysine N-methyltransferase MLL3, Histone-lysine N-methyltransferase MLL4, Histone-lysine N-methyltransferase MLL5, Histone-lysine N-methyltransferase NSD3, Histone-lysine N-methyltransferase PRDM9, Histone-lysine N-methyltransferase SETD1A, Histone-lysine N-methyltransferase SETD1B, Histone-lysine N-methyltransferase SETD2, Histone-lysine N-methyltransferase SETD7, Histone-lysine N-methyltransferase SETD8, Histone-lysine N-methyltransferase SETDB1, Histone-lysine N-methyltransferase SETDB2, Histone-lysine N-methyltransferase SETMAR, Histone-lysine N-methyltransferase SUV39H1, Histone-lysine N-methyltransferase SUV39H2, Histone-lysine N-methyltransferase SUV420H1, Histone-lysine N-methyltransferase SUV420H2, Histone-lysine N-methyltransferase, H3 lysine-36 and H4 lysine-20 specific, Histone-lysine N-methyltransferase, H3 lysine-79 specific, Histone-lysine N-methyltransferase, H3 lysine-9 specific 3, Histone-lysine N-methyltransferase, H3 lysine-9 specific 5, N-lysine methyltransferase SMYD2, PR domain zinc finger protein 2, Probable histone-lysine N-methyltransferase ASH1L, Probable histone-lysine N-methyltransferase NSD2, SET and MYND domain-containing protein 3